4 Sourcing data
4.1 Introduction
Welcome to your fourth week of Introduction to Quantitative Research Methods. This week we will introduce you to sourcing and preparing data from official sources. For the tutorial we will apply what we have learnt over the past weeks onto a new data set.
This week is structured by 4 short videos, practical material that you need to work through in preparation for Thursday’s seminar, and a seminar task that you need to do in preparation for Thursday’s seminar.
Let’s do it.
Video: Introduction W4
[Lecture slides] [Watch on MS stream]4.1.1 Reading list
Please find the reading list for this week below. We strongly recommend that you read the core reading material before you continue with the rest of this week’s material.
Core reading
- Wickham, 2014, Tidy data, Journal of Statistical Software 59(10). [Link]
Supplementary reading
- Zhang et al., 2010, Data preparation for data mining. Applied Artificial Intelligence 17(5-6): 375-381. [Link]
4.1.2 Q&A session
This week there is NO live Q&A session scheduled. Please post any questions you have on the POLS0008 Forum or ask them to your seminar leader during your live seminar on Thursday.
4.2 Sourcing data
Over the past weeks we have predominantly worked with two data sets that we provided for you: ambulance-assault.csv and census-historic-population-borough.csv. Although these data sets are very useful to give you an introduction to Quantitative Research methods, in real life data you will have to source your own data. In the case of the social sciences, you often can find socio-economic data on the websites of national statistical authorities such as the Office for National Statistics, United States Census Bureau, Statistics South Africa, or the National Bureau of Statistics of China. Also large international institutions like World Bank and Unicef compile socio-economic data sets and make these available for research purposes. No matter where you will be getting your data from, however, you will have to know how to download and prepare these data sets in such a way that you can work with them in R to conduct your analysis.
Video: Sourcing data I
[Lecture slides] [Watch on MS stream]Within the United Kingdom, the Office for National Statistics (ONS) is the largest producer of official statistics. ONS is responsible for collecting and publishing statistics related to the economy, population and society at national, regional and local levels. They are also responsible for conducting the census in England and Wales every 10 years (the census for Scotland and Northern Ireland are conducted by the National Records of Scotland and the Northern Ireland Statistics and Research Agency, respectively). Today we will be downloading a data set from ONS, prepare the data set as a csv file, and read our freshly created csv file into R.
Figure 4.1: The website of the Office for National Statistics.
Because the population data in the census-historic-population-borough.csv we have been working with so far only goes as far as 2011, we will try to get some more recent population estimates on the ‘usual resident population’. Every year, ONS releases a new set of Middle Super Output Area mid-year population estimates. Currently, the latest available data set is that of mid-2019 and this is the data set that we are now going to download and prepare.
Note
The mid-year population estimates that we will be downloading are provided by ONS at the Middle Super Output Area (MSOA) level. MSOAs are one of the many administrative geographies that the ONS uses for reporting their small area statistics. An administrative geography is a way of dividing the country into smaller sub-divisions or areas that correspond with the area of responsibility of local authorities and government bodies. These geographies are updated as populations evolve and as a result, the boundaries of the administrative geographies are subject to either periodic or occasional change. The UK has quite a complex administrative geography, particularly due to having several countries within one overriding administration and then multiple ways of dividing the countries according to specific applications.
To download the data set, you need to take the following steps:
| Step | Action |
|---|---|
| 1 | Navigate to the download page of the Middle Super Output Area population estimates: [Link] |
| 2 | Download the file Mid-2019: SAPE22DT14 to your computer. |
| 3 | Because the file that you have now downloaded is a zip file, we first need to extract the file before we can use it. To unzip the file you can use the built-in functionality of your computer’s operating system. For Windows: right click on the zip file, select Extract All, and then follow the instructions. For Mac OS: double-click the zip file. |
| 4 | Open the file in Microsoft Excel or any other spreadsheet software. Please note that the instructions provided below use only cover Microsoft Excel. |
| 5 | Once opened your file should look similar as the screenshot in Figure 4.2. |
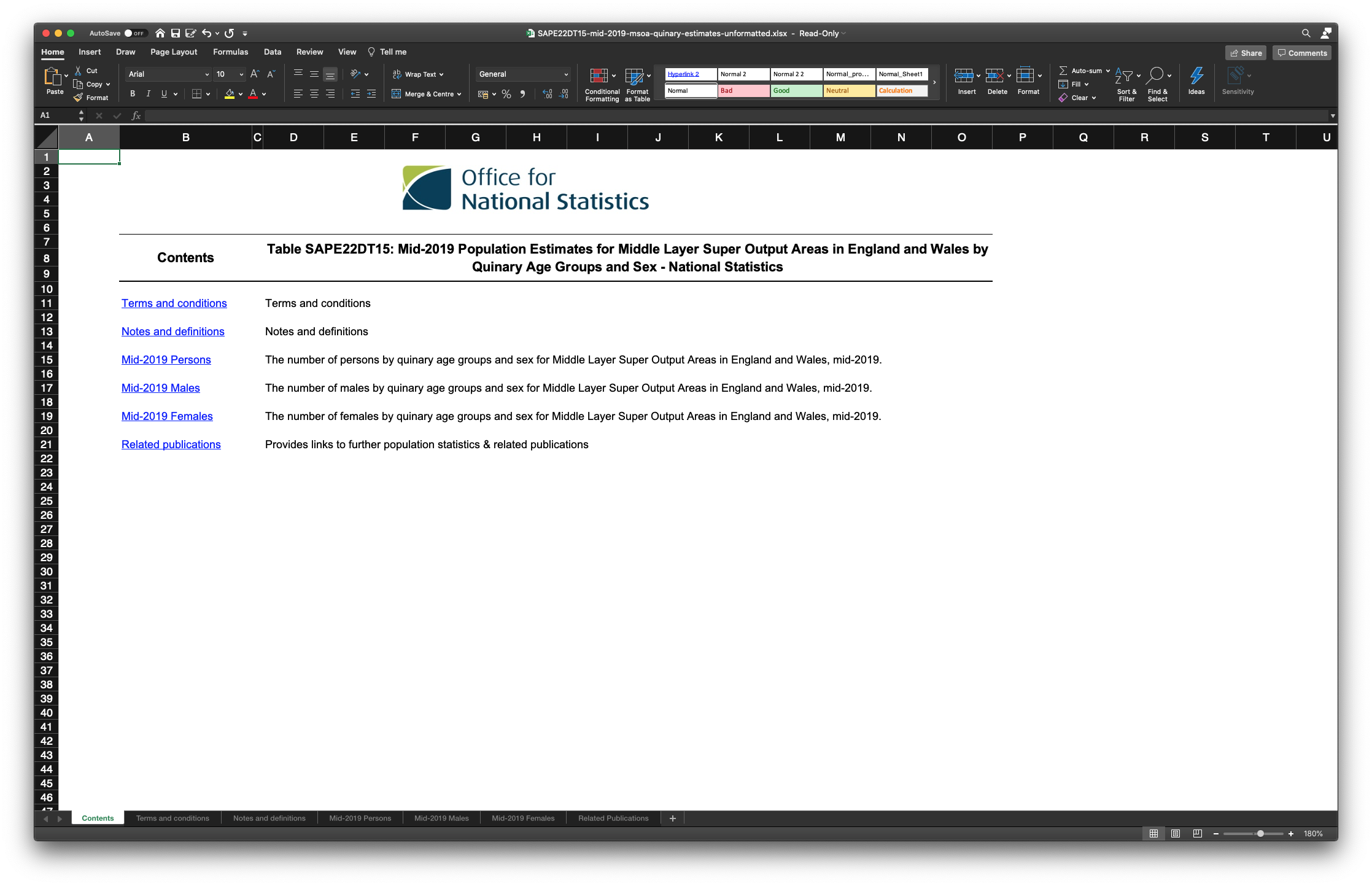
Figure 4.2: The Mid-2019: SAPE22DT15 file that we downloaded.
The file probably does not look exactly like you thought it would because we do not directly see any data! In fact, ONS has put the data on different tabs. Some of these tabs contain the actual data, whilst others contain some meta-data and notes and definitions. The data that we want to use is found on the Mid-2019 Persons tab: the total number of people living in each MSOA. The problem we have now, however, is that the data as it is right now cannot be read into R without causing us lots of problems down the road. So even though we have the data we want to work with at our finger tips, we are not yet ready to read our data set into R! So, in video below, we will show you how to create a csv from the data that we want to use.
Video: Sourcing data II
[Lecture slides] [Watch on MS stream]Now you have seen how to prepare your csv, it is time to do it yourselves. Just as a small recap, these are the steps that you need to take to turn the downloaded data into a csv:
| Step | Action |
|---|---|
| 1 | Open the downloaded file in Microsoft Excel and activate the Mid-2019 Persons tab. |
| 2 | Highlight the columns: MSOA Code, MSOA Name, LA Code (2019 boundaries), LA name (2019 boundaries), LA Code (2020 boundaries), LA name (2020 boundaries), and All Ages. Do make sure that you do not include any of the whitespace or empty rows. |
| 3 | Scroll down all the way to the bottom of the file. Now hold down the shift button and click on the last value in the All Ages column. This value should be 9,711. All data should now be selected. |
| 4 | Now all the data that we need are selected we can copy them by right clicking and in the context menu opting for the copy option. Of course, you can also simply use control + c (Windows) or command + c (Mac OS). |
| 5 | Open a new, empty spreadsheet and paste the copied data into this new, empty spreadsheet. You can paste your copied data by right clicking on the first cell (A1) and in the context menu opting for the paste option. Of course, you can also simply use control + v (Windows) or command + v (Mac OS). |
| 6 | Conduct a visual check to make sure that you copied all the data. |
| 7 | Conduct a visual check to make sure that you did not copy any additional data. |
| 8 | Remove all formatting and make sure that empty columns are indeed empty. |
| 9 | Save this file as a midyear2019.csv. Make sure that you select csv as your file format (e.g. CSV UTF8 (Comma-delimited) (.csv)). |
| 10 | Inspect your data in a text editor such as Wordpad or Textedit to make sure the file you created is indeed a comma-separated file. |
Now, for the moment of truth: let’s try and see if we can load our data into R! Of course, you will first need to upload your csv file to RStudio Server and set your working directory so that R can find the file - but you should be able to do that without any problems by now.
# load csv file from working directory
midyear <- read.csv('midyear2019.csv')## MSOA.Code MSOA.Name LA.Code..2019.boundaries.
## 1 E02002483 Hartlepool 001 E06000001
## 2 E02002484 Hartlepool 002 E06000001
## 3 E02002485 Hartlepool 003 E06000001
## 4 E02002487 Hartlepool 005 E06000001
## LA.name..2019.boundaries. LA.Code..2020.boundaries.
## 1 Hartlepool E06000001
## 2 Hartlepool E06000001
## 3 Hartlepool E06000001
## 4 Hartlepool E06000001
## LA.name..2020.boundaries. All.Ages
## 1 Hartlepool 10 261
## 2 Hartlepool 10 449
## 3 Hartlepool 8 185
## 4 Hartlepool 5 277
## [ reached 'max' / getOption("max.print") -- omitted 7197 rows ]Questions
- Inspect the column names. Why do you think that they slightly differ from the ones that we saw in Microsoft Excel?
- How many rows does the
midyeardataframe have? - How many columns does the
midyeardataframe have?
The column names may seem a little complicated but in fact simply refer to some of the administrative geographies that the ONS uses for their statistics. Don’t worry if you do not fully understand these as they are notoriously complicated and confusing!
| Column heading | Full name | Description |
|---|---|---|
| MSOA.Code | MSOA Code | Middle Super Output Areas are one of the many administrative geographies that the ONS uses for reporting their small area statistics. These codes are used as a quick way of referring to them from official data sources. |
| MSOA.Name | MSOA Name | Name of the MSOA. |
| LA.Code..2019.boundaries | Local Authority District codes 2019 | Local Authority Districts are a subnational division used for the purposes of local government. Each MSOA belongs to one Local Authority District, however, between years the boundaries of these Local Authority Districts sometimes change. These are the codes of the Local Authority District to which the MSOA belonged to in 2019. |
| LA.name..2019.boundaries | Local Authority District names 2019 | Name of the Local Authority District to which the MSOA belonged to in 2019. |
| LA.Code..2020.boundaries | Local Authority District codes 2020 | Code of the Local Authority District to which the MSOA belonged to in 2020 |
| LA.name..2020.boundaries | Local Authority District names 2020 | Name of the Local Authority District to which the MSOA belonged to in 2019. |
| All.Ages | Total number of people | The total number of people that is estimated to live in the MSOA mid-2019. |
Now we at least have an idea about what our column names mean, we can rename them so that they are a little more intelligible and easier to work with in a later stage.
# rename columns
names(midyear) <- c('msoa_code','msoa_name','lad19_code','lad19_name','lad20_code','lad20_name','pop19')
# inspect
names(midyear)## [1] "msoa_code" "msoa_name" "lad19_code" "lad19_name" "lad20_code"
## [6] "lad20_name" "pop19"# write to new csv
write.csv(midyear, 'midyear2019_v2.csv', row.names=FALSE)Great. This is all looking very good but we are not completely there yet. Now we sourced our data and managed to successfully read the data set into R, we will need to do conduct some final preparations so that our data is analysis ready.
Recap
In this section you have learnt how to:
- Download data from the Office for National Statistics.
- Select the data that you need and save these into a
csvfile. - Load the
csvfile that you created into R.
4.3 Preparing data
Even though the data set that we downloaded came from an official national statistical authority, the data is not directly ready for analysis. In fact, the vast majority of the data you will find in the public domain (or private domain for that matter) will be dirty data. With dirty data we mean data that needs some level of pre-processing, cleaning, and linkage before you can use it for your analysis. In the following, you will learn a consistent way to structure your data in R: tidy data. Tidy data, as formalised by R expert Hadley Wickham in his contribution to the Journal of Statistical Software is not only very much at the core of the tidyverse R package that we will introduce you to, but also of general importance when organising your data.
Video: Preparing data
[Lecture slides] [Watch on MS stream]As you know by now, your basic R functionality (the base package) can be extended by installing additional packages. These packages are the fundamental units of reproducible R code and include functions, documentation, and sample data. Last week, we introduced you to the ggplot2 package: a general scheme for data visualisation that breaks up graphs into semantic components such as scales and layers. Today we will introduce you to the tidyverse package. This package is specifically created to make working with data, including data cleaning and data preparation, easier. Let’s start by installing the tidyverse package using the install_packages() function in the same way as we installed the ggplot2 package last week.
# install the tidyverse
install.packages('tidyverse')Note
The tidyverse package is in fact a collection of packages that are specifically designed for data science tasks. Where in many cases different packages work all slightly differently, all packages of the tidyverse share the underlying design philosophy, grammar, and data structures. The ggplot2 package that you worked with last week is actually one of the core package of the tidyverse. This also means that if you load the tidyverse package through library(tidyverse) you directly have access to all the functions that are part of the ggplot2 package and you do not have to load the ggplot2 package seperately. Because the tidyverse consists of multiple packages, it may take a little while before everything is installed so be patient! For more information on tidyverse, have a look at https://www.tidyverse.org/.
If your installation was successful, you can now load the tidyverse as follows:
# load the tidyverse
library(tidyverse)## ── Attaching packages ─────────────────────────────────────────────── tidyverse 1.3.0 ──## ✓ tibble 3.0.4 ✓ dplyr 1.0.3
## ✓ tidyr 1.1.2 ✓ stringr 1.4.0
## ✓ readr 1.4.0 ✓ forcats 0.5.0
## ✓ purrr 0.3.4## ── Conflicts ────────────────────────────────────────────────── tidyverse_conflicts() ──
## x dplyr::filter() masks stats::filter()
## x dplyr::lag() masks stats::lag()
Figure 4.3: Loading the tidyverse.
Note
After loading the tidyverse, you may get a short information messages to inform you which packages are attached to your R session and whether there are any conflicting functions. This simply means that there are functions that are named the same across the packages that you have loaded. For instance, when you load the tidyverse in a fresh R session, the tidyverse will tell you that two functions from the dplyr package (which is an integral part of the tidyverse package) mask the functionality of two functions of the stats package (which is part of base R). This simply means that if you now type in filter() you will get the dplyr functionality and not the functionality of the stats. This is very important information as both these functions do something completely different! You can still use the filter() function from the stats package but then you have to explicitly tell R that you want to use the function called filter() within the stats package: stats::filter().
Now that we have loaded the tidyverse we can continue with our data preparation. We will start by reading in the csv file that we created earlier, however, we will use the function read_csv() from the tidyverse instead of the function read.csv() from base R. The function read_csv returns a so-called tibble. Tibbles are data frames, but they tweak some older behaviours to make life a little easier.
# load csv file from working directory
midyear <- read_csv('midyear2019_v2.csv')# inspect
midyear## # A tibble: 7,201 x 7
## msoa_code msoa_name lad19_code lad19_name lad20_code lad20_name pop19
## <chr> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 E02002483 Hartlepool … E06000001 Hartlepool E06000001 Hartlepool 10 2…
## 2 E02002484 Hartlepool … E06000001 Hartlepool E06000001 Hartlepool 10 4…
## 3 E02002485 Hartlepool … E06000001 Hartlepool E06000001 Hartlepool 8 185
## 4 E02002487 Hartlepool … E06000001 Hartlepool E06000001 Hartlepool 5 277
## 5 E02002488 Hartlepool … E06000001 Hartlepool E06000001 Hartlepool 5 848
## 6 E02002489 Hartlepool … E06000001 Hartlepool E06000001 Hartlepool 7 730
## 7 E02002490 Hartlepool … E06000001 Hartlepool E06000001 Hartlepool 6 214
## 8 E02002491 Hartlepool … E06000001 Hartlepool E06000001 Hartlepool 6 454
## 9 E02002492 Hartlepool … E06000001 Hartlepool E06000001 Hartlepool 6 887
## 10 E02002493 Hartlepool … E06000001 Hartlepool E06000001 Hartlepool 6 702
## # … with 7,191 more rowsQuestions
- Do you notice a difference between how
baseR prints a data frame and how atibblegets printed?
Let’s have a better look at our data particularly the data types. We can can do this through the str() function:
# inspect
str(midyear)## tibble [7,201 × 7] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
## $ msoa_code : chr [1:7201] "E02002483" "E02002484" "E02002485" "E02002487" ...
## $ msoa_name : chr [1:7201] "Hartlepool 001" "Hartlepool 002" "Hartlepool 003" "Hartlepool 005" ...
## $ lad19_code: chr [1:7201] "E06000001" "E06000001" "E06000001" "E06000001" ...
## $ lad19_name: chr [1:7201] "Hartlepool" "Hartlepool" "Hartlepool" "Hartlepool" ...
## $ lad20_code: chr [1:7201] "E06000001" "E06000001" "E06000001" "E06000001" ...
## $ lad20_name: chr [1:7201] "Hartlepool" "Hartlepool" "Hartlepool" "Hartlepool" ...
## $ pop19 : chr [1:7201] "10 261" "10 449" "8 185" "5 277" ...
## - attr(*, "spec")=
## .. cols(
## .. msoa_code = col_character(),
## .. msoa_name = col_character(),
## .. lad19_code = col_character(),
## .. lad19_name = col_character(),
## .. lad20_code = col_character(),
## .. lad20_name = col_character(),
## .. pop19 = col_character()
## .. )The output of the str() function tells us that every column in our data frame (tibble) is of type “character”, which comes down to a variable that contains textual data. What is interesting, obviously, is that also our pop19 variable is considered a character variable. The reason for this is that in the original data white space is used as thousands separator (i.e. 11 000 instead of 11000 or 11,000). Does it matter? Well, let’s try to calculate the mean and median of our pop19 variable:
# mean
mean(midyear$pop19)## Warning in mean.default(midyear$pop19): argument is not numeric or logical:
## returning NA## [1] NA# median
median(midyear$pop19)## [1] "7 251"Note
Depending on computer settings and the version of Microsoft Excel or spreadsheet software that you use, in some case you could get a numeric variable (e.g. double) instead of a character variable for your pop19 variable. If so: you are lucky and do not need to undertake the steps below to turn this character column into a numeric column. (This will also mean that at this stage your results will differ from the results shown here.) Just continue with the tutorial by carefully reading through the steps that you would have needed to take if indeed your results would have been the same as shown here.
As you can see, this is not working perfectly for the simple fact that the the mean() function requires a numeric variable as its input! The median() does return a value, however, this is not per se the median that we are interested in as the sorting of the data is now done alphabetically: see for your self what happens when using the sort() function on the pop19 variable. Fortunately, there are some functions in the dplyr package, which is part of the tidyverse, that will help us further cleaning and preparing our data. Some of the most important and useful functions are:
| Package | Function | Use to |
|---|---|---|
| dplyr | select | select columns |
| dplyr | filter | select rows |
| dplyr | mutate | transform or recode variables |
| dplyr | summarise | summarise data |
| dplyr | group by | group data into subgroups for further processing |
Note
Remember that when you encounter a function in a piece of R code that you have not seen before and you are wondering what it does that you can get access the documentation through ?name_of_function, e.g. ?mutate. For almost any R package, the documentation contains a list of arguments that the function takes, in which format the functions expects these arguments, as well as a set of usage examples.
Let’s have a look at two of the dplyr functions: select() and filter().
# select columns with the select function
# note that within dplyr we not use quotation marks to refer to columns
midyear_sel <- select(midyear,msoa_code,msoa_name,lad20_code,lad20_name,pop19)
# inspect
midyear_sel## # A tibble: 7,201 x 5
## msoa_code msoa_name lad20_code lad20_name pop19
## <chr> <chr> <chr> <chr> <chr>
## 1 E02002483 Hartlepool 001 E06000001 Hartlepool 10 261
## 2 E02002484 Hartlepool 002 E06000001 Hartlepool 10 449
## 3 E02002485 Hartlepool 003 E06000001 Hartlepool 8 185
## 4 E02002487 Hartlepool 005 E06000001 Hartlepool 5 277
## 5 E02002488 Hartlepool 006 E06000001 Hartlepool 5 848
## 6 E02002489 Hartlepool 007 E06000001 Hartlepool 7 730
## 7 E02002490 Hartlepool 008 E06000001 Hartlepool 6 214
## 8 E02002491 Hartlepool 009 E06000001 Hartlepool 6 454
## 9 E02002492 Hartlepool 010 E06000001 Hartlepool 6 887
## 10 E02002493 Hartlepool 011 E06000001 Hartlepool 6 702
## # … with 7,191 more rows# filter rows with the filter function
# note that within dplyr we not use quotation marks to refer to columns
midyear_fil <- filter(midyear,lad20_name=='Leeds')
# inspect
midyear_fil## # A tibble: 107 x 7
## msoa_code msoa_name lad19_code lad19_name lad20_code lad20_name pop19
## <chr> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 E02002330 Leeds 001 E08000035 Leeds E08000035 Leeds 6 572
## 2 E02002331 Leeds 002 E08000035 Leeds E08000035 Leeds 7 029
## 3 E02002332 Leeds 003 E08000035 Leeds E08000035 Leeds 6 143
## 4 E02002333 Leeds 004 E08000035 Leeds E08000035 Leeds 7 637
## 5 E02002334 Leeds 005 E08000035 Leeds E08000035 Leeds 7 110
## 6 E02002335 Leeds 006 E08000035 Leeds E08000035 Leeds 7 500
## 7 E02002336 Leeds 007 E08000035 Leeds E08000035 Leeds 6 596
## 8 E02002337 Leeds 008 E08000035 Leeds E08000035 Leeds 7 388
## 9 E02002338 Leeds 009 E08000035 Leeds E08000035 Leeds 6 817
## 10 E02002339 Leeds 010 E08000035 Leeds E08000035 Leeds 5 351
## # … with 97 more rowsQuestions
- How can we get the same data frame with selected columns (
midyear_sel) using thebaseR syntax we have been using in previous weeks? - How can we get the same data frame with filtered rows (
midyear_fil) using thebaseR syntax we have been using in previous weeks?
With the mutate() function we can easily create new columns, so let’s try this function to see if we can create a new variable that contains all the data from pop19 in a numeric format.
# create a new variable named pop19_no space
# by replacing all white space with nothing using the "str_replace_all" function
midyear <- mutate(midyear, pop19_nospace=str_replace_all(pop19, pattern=' ', repl=''))
# inspect
midyear## # A tibble: 7,201 x 8
## msoa_code msoa_name lad19_code lad19_name lad20_code lad20_name pop19
## <chr> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 E02002483 Hartlepo… E06000001 Hartlepool E06000001 Hartlepool 10 2…
## 2 E02002484 Hartlepo… E06000001 Hartlepool E06000001 Hartlepool 10 4…
## 3 E02002485 Hartlepo… E06000001 Hartlepool E06000001 Hartlepool 8 185
## 4 E02002487 Hartlepo… E06000001 Hartlepool E06000001 Hartlepool 5 277
## 5 E02002488 Hartlepo… E06000001 Hartlepool E06000001 Hartlepool 5 848
## 6 E02002489 Hartlepo… E06000001 Hartlepool E06000001 Hartlepool 7 730
## 7 E02002490 Hartlepo… E06000001 Hartlepool E06000001 Hartlepool 6 214
## 8 E02002491 Hartlepo… E06000001 Hartlepool E06000001 Hartlepool 6 454
## 9 E02002492 Hartlepo… E06000001 Hartlepool E06000001 Hartlepool 6 887
## 10 E02002493 Hartlepo… E06000001 Hartlepool E06000001 Hartlepool 6 702
## # … with 7,191 more rows, and 1 more variable: pop19_nospace <chr># create a new variable named pop19_numeric
# by transorming the pop19_nospace variable using the "as.numeric" function
midyear <- mutate(midyear,pop19_numeric=as.numeric(pop19_nospace))
# inspect
midyear## # A tibble: 7,201 x 9
## msoa_code msoa_name lad19_code lad19_name lad20_code lad20_name pop19
## <chr> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 E02002483 Hartlepo… E06000001 Hartlepool E06000001 Hartlepool 10 2…
## 2 E02002484 Hartlepo… E06000001 Hartlepool E06000001 Hartlepool 10 4…
## 3 E02002485 Hartlepo… E06000001 Hartlepool E06000001 Hartlepool 8 185
## 4 E02002487 Hartlepo… E06000001 Hartlepool E06000001 Hartlepool 5 277
## 5 E02002488 Hartlepo… E06000001 Hartlepool E06000001 Hartlepool 5 848
## 6 E02002489 Hartlepo… E06000001 Hartlepool E06000001 Hartlepool 7 730
## 7 E02002490 Hartlepo… E06000001 Hartlepool E06000001 Hartlepool 6 214
## 8 E02002491 Hartlepo… E06000001 Hartlepool E06000001 Hartlepool 6 454
## 9 E02002492 Hartlepo… E06000001 Hartlepool E06000001 Hartlepool 6 887
## 10 E02002493 Hartlepo… E06000001 Hartlepool E06000001 Hartlepool 6 702
## # … with 7,191 more rows, and 2 more variables: pop19_nospace <chr>,
## # pop19_numeric <dbl># inspect
str(midyear)## tibble [7,201 × 9] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
## $ msoa_code : chr [1:7201] "E02002483" "E02002484" "E02002485" "E02002487" ...
## $ msoa_name : chr [1:7201] "Hartlepool 001" "Hartlepool 002" "Hartlepool 003" "Hartlepool 005" ...
## $ lad19_code : chr [1:7201] "E06000001" "E06000001" "E06000001" "E06000001" ...
## $ lad19_name : chr [1:7201] "Hartlepool" "Hartlepool" "Hartlepool" "Hartlepool" ...
## $ lad20_code : chr [1:7201] "E06000001" "E06000001" "E06000001" "E06000001" ...
## $ lad20_name : chr [1:7201] "Hartlepool" "Hartlepool" "Hartlepool" "Hartlepool" ...
## $ pop19 : chr [1:7201] "10 261" "10 449" "8 185" "5 277" ...
## $ pop19_nospace: chr [1:7201] "10261" "10449" "8185" "5277" ...
## $ pop19_numeric: num [1:7201] 10261 10449 8185 5277 5848 ...
## - attr(*, "spec")=
## .. cols(
## .. msoa_code = col_character(),
## .. msoa_name = col_character(),
## .. lad19_code = col_character(),
## .. lad19_name = col_character(),
## .. lad20_code = col_character(),
## .. lad20_name = col_character(),
## .. pop19 = col_character()
## .. )Note the information for pop19_numeric: str indicates that the column is now of type num (i.e. numeric). This is much better. We can now try to calculate the median and the mean again:
# calculate the mean of the pop19_numeric variable
mean(midyear$pop19_numeric)## [1] 8254.387# calculate the median of the pop19_numeric variable
median(midyear$pop19_numeric)## [1] 7955Everything seems to be working fine now and our data is now finally ready for analysis, so it is probably wise to save’s save it.
# write to new csv
write.csv(midyear, 'midyear2019_clean.csv', row.names=FALSE)Recap
In this section you have learnt how to:
- Transform your data frame to a tibble.
- Use some of the functionality of the
tidyverseto prepare your data set and make it suitable for analysis.
4.4 Seminar
Please find the seminar task and seminar questions for this week’s seminar below.
Note
Please make sure that you have executed the seminar task and have answered the seminar questions before the seminar!
Seminar task
Of course, we did not go through all the trouble of downloading and preparing this file without using it. Use the cleaned version of the midyear data set that we just created to:
- Create a new object / data set that only contains data for the Local Authority District (2020 boundaries) of Manchester.
- Create a new object / data set that only contains data for the Local Authority District (2020 boundaries) of Birmingham.
For both new objects:
- Calculate the mean, median, and standard deviation of the pop19_numeric variable.
- Create a boxplot of the pop19_numeric variable using the
ggplot2package. - Create a histogram of the pop19_numeric variable using the
ggplot2package. Select a bin width that you think is appropriate.
Seminar questions
- Compare the results of the descriptive statistics you have calculated for your Birmingham object / data set with the results of the descriptive statistics you have calculated for you Manchester data set. What do these descriptive statistics tell you?
- Why did you select the bin width that you used for creating your histograms?
- Compare the histograms that you created for your Birmingham object / data set and the Manchester data set, what can you tell about the population distribution of both Local Authority Districts?
Additional tasks
If you found these assignments and questions relatively easy and want an assignment that is a little more realistic, try to do the following:
- Download the mid-year population estimates for 2018 to your computer (
Mid-2018: SAPE21DT15 (unformatted)). Make sure that you download the file with exactly this name: it is listed under the “supporting files you may find useful” heading. - Prepare a
csvfile of the first three columns of theMid-2018 Peronstab and import into R. - Give the columns the following names: msoa_code, msoa_name, pop18.
- Take the necessary steps in R to make sure that all numeric variables are also recognised by R as numeric variables.
- Join the 2018 and 2019 midyear population data set by using the
left join()function from thetidyverse()package, e.g. something like:midyear <- left_join(midyear2018, midyear2019, by=c('msoa_code'='msoa_code'). - Calculate the population change between 2018 and 2019.
- Create a histogram of the population change.
- Which MSOA has the largest decline in population between 2018 and 2019? Which MSOA had the largest increase in population between 2018 and 2019?
Seminar link
Seminars for all groups take place on Thursday morning. You can find the Zoom link to your seminar group on Moodle.
4.5 Before you leave
Save your R script by pressing the Save button in the script window. That is it for this week!